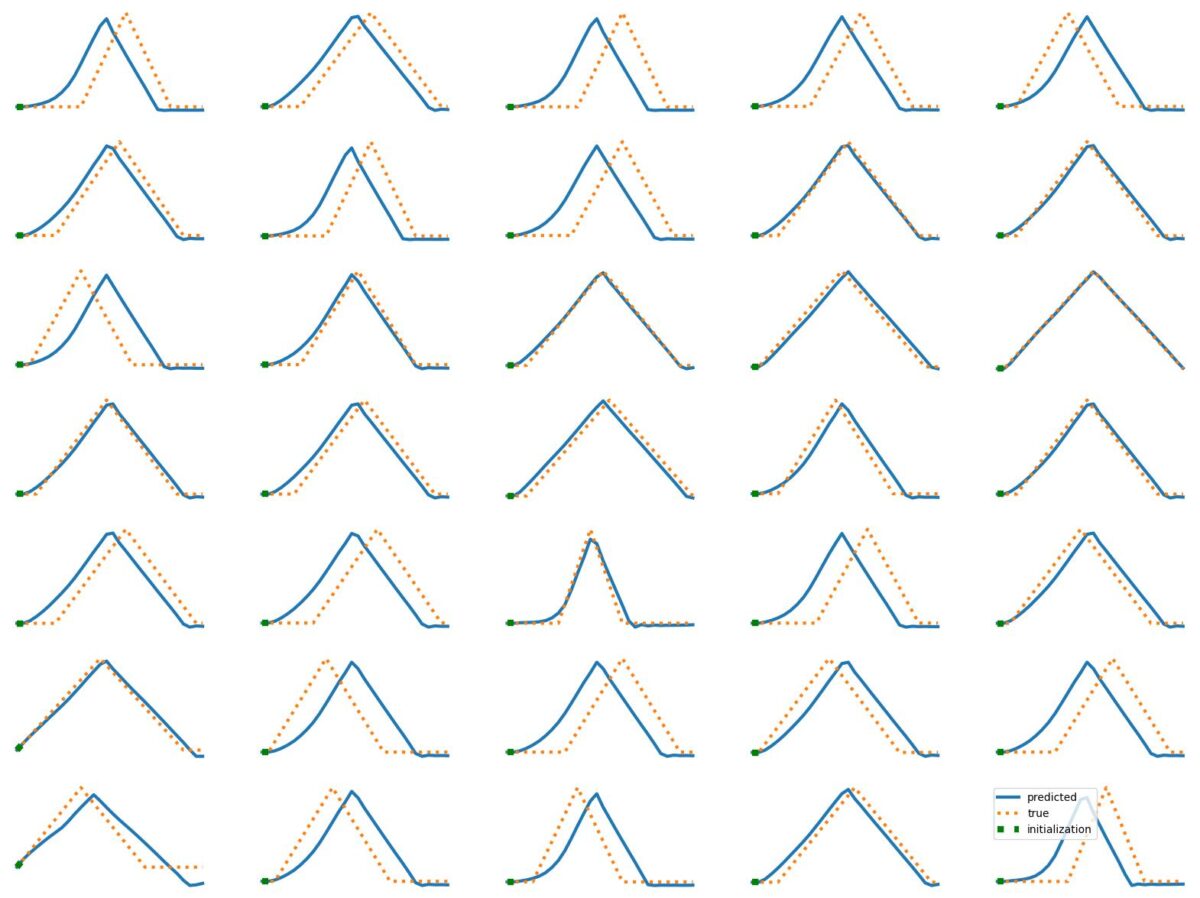
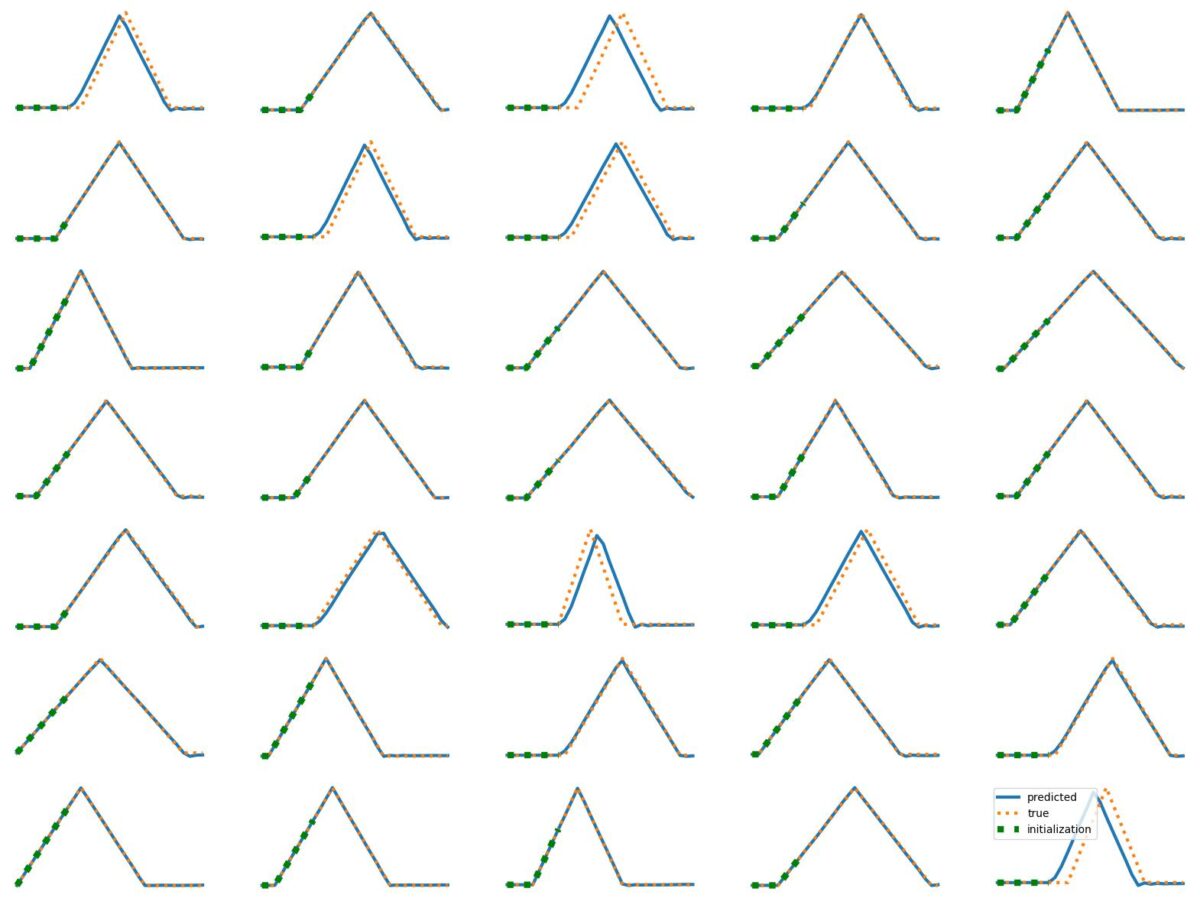
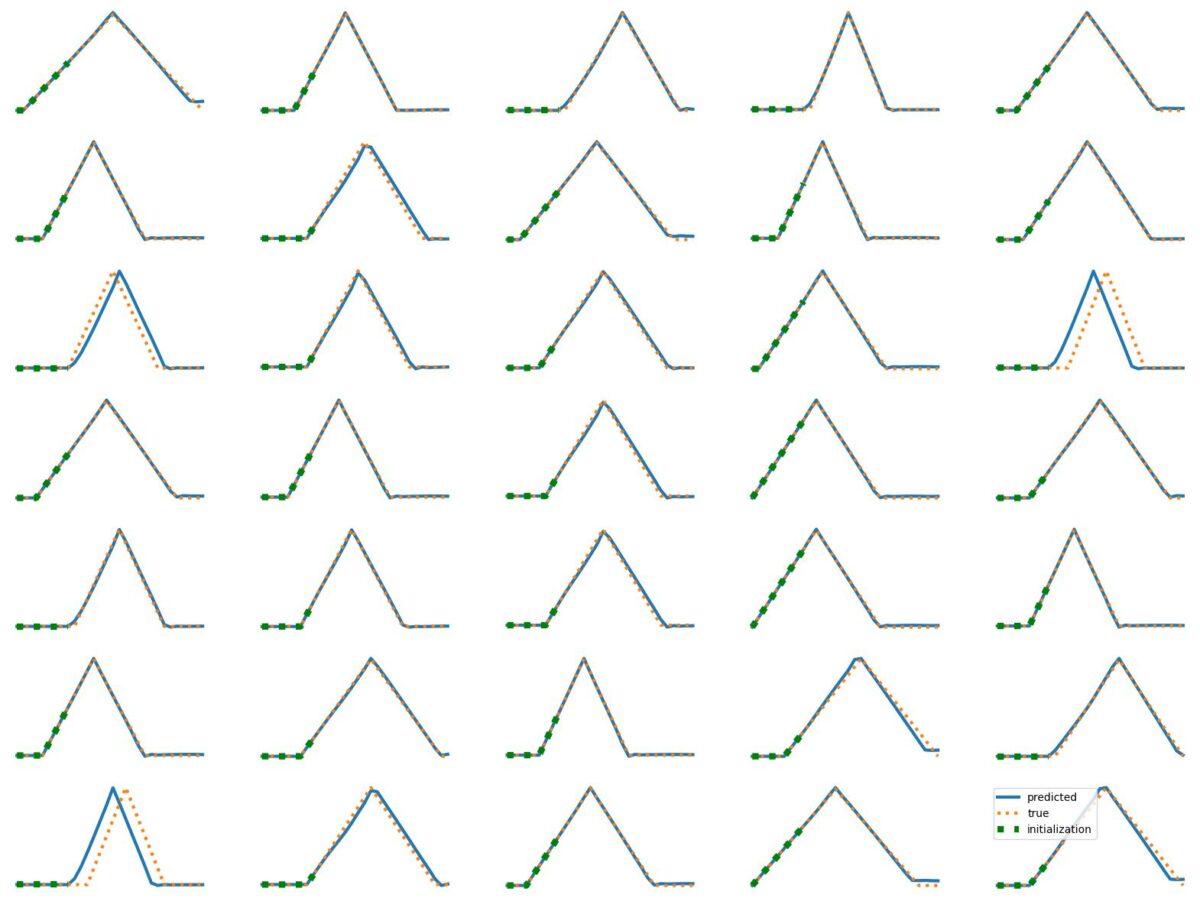
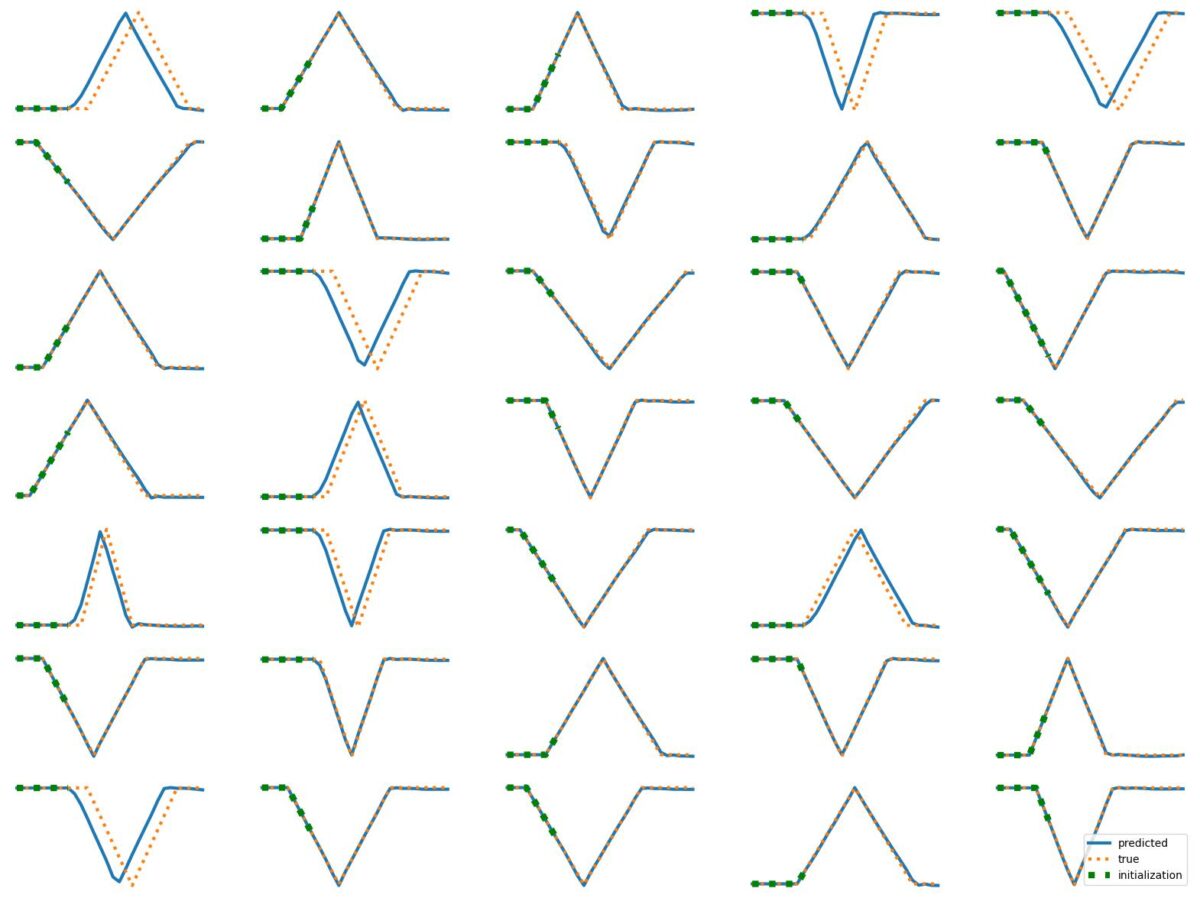
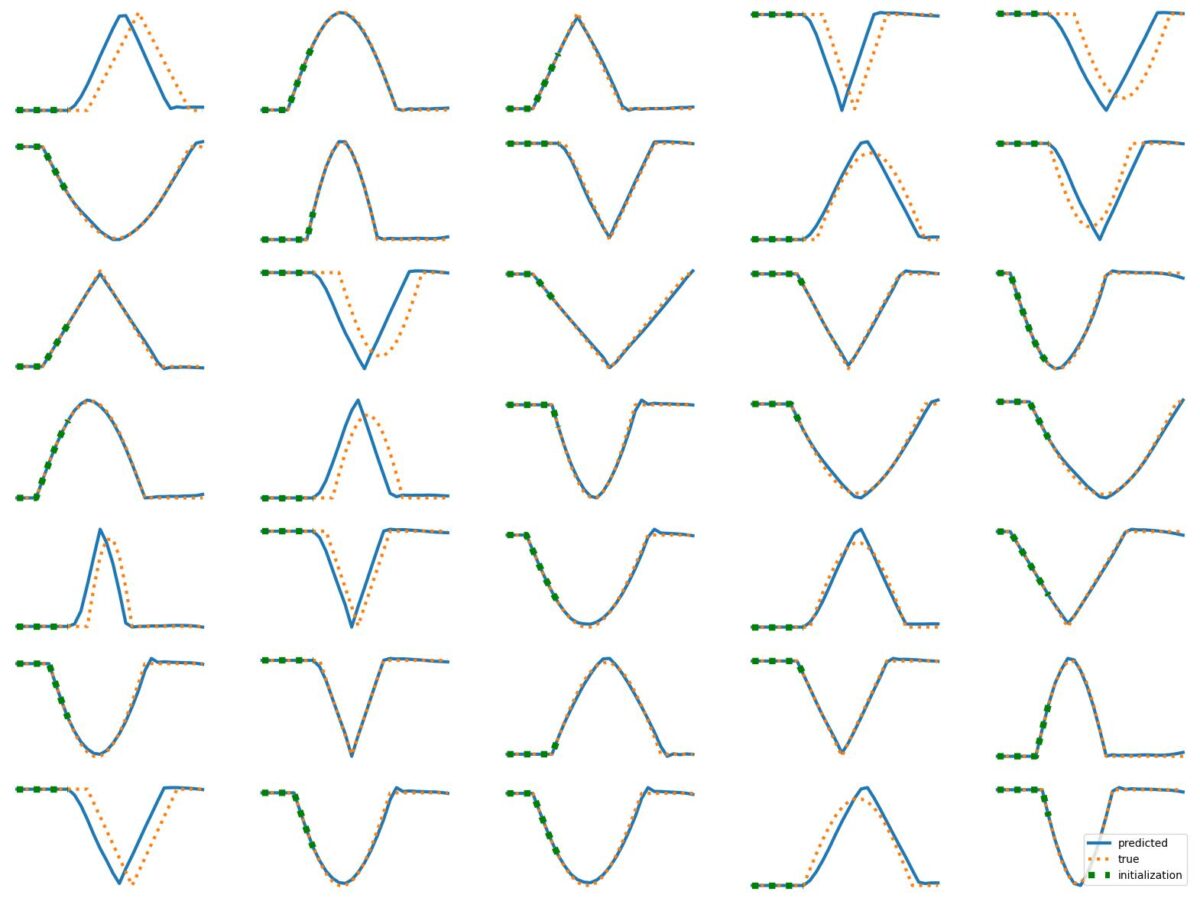
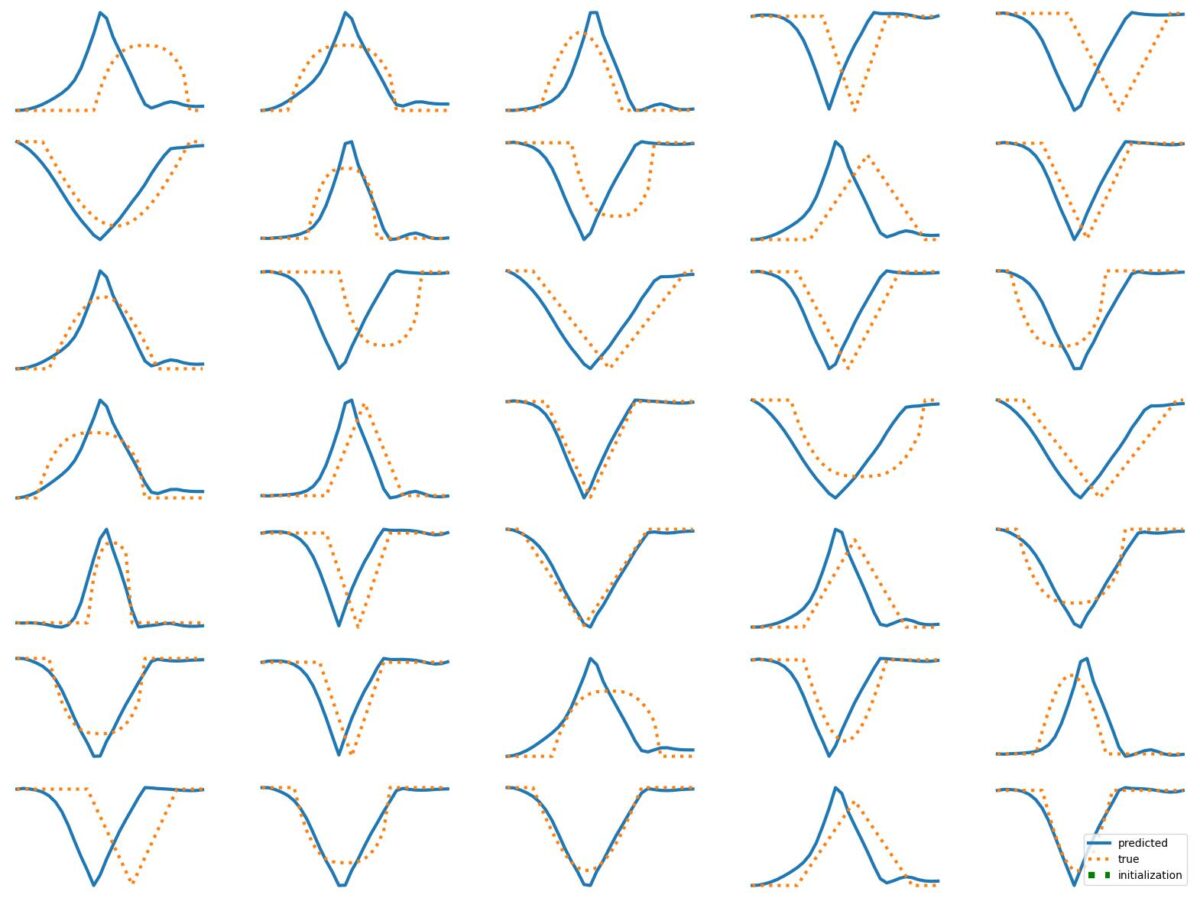
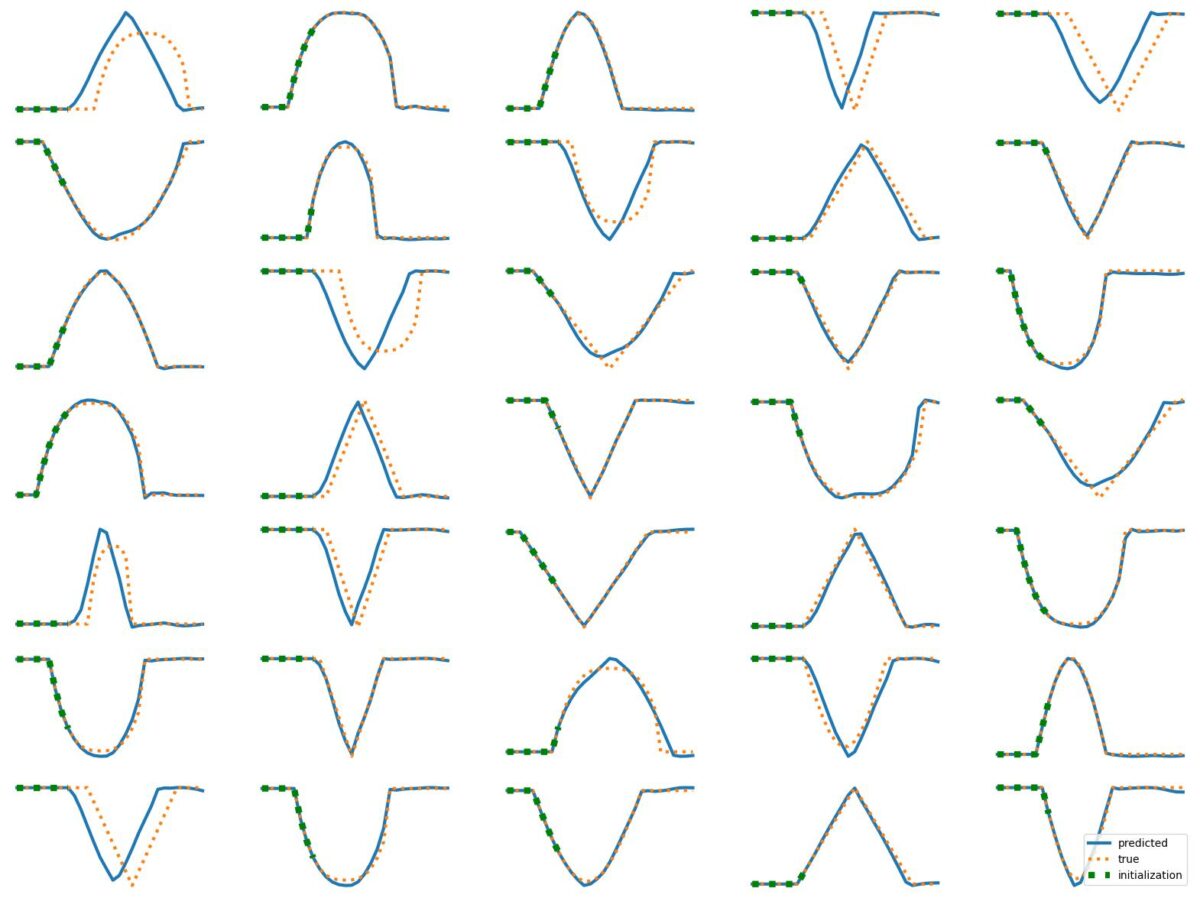
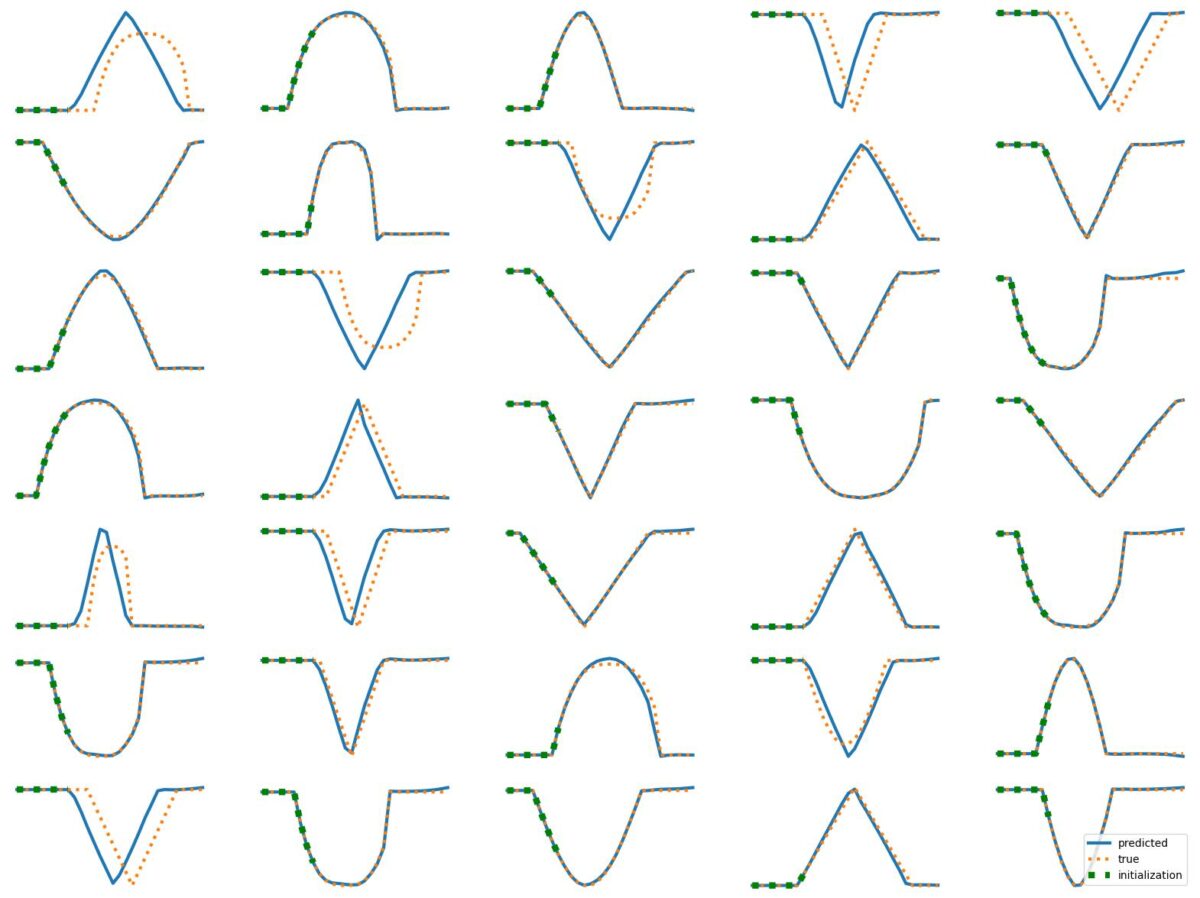
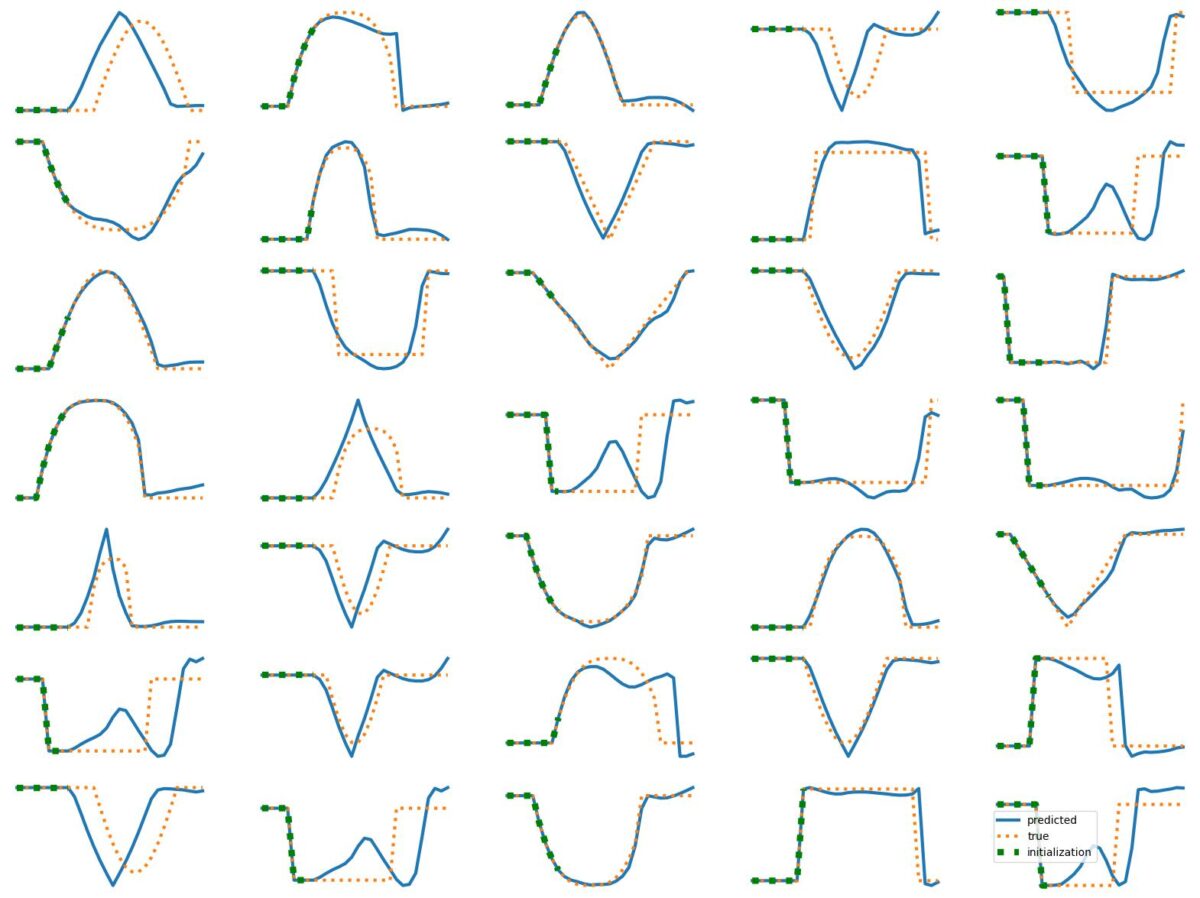
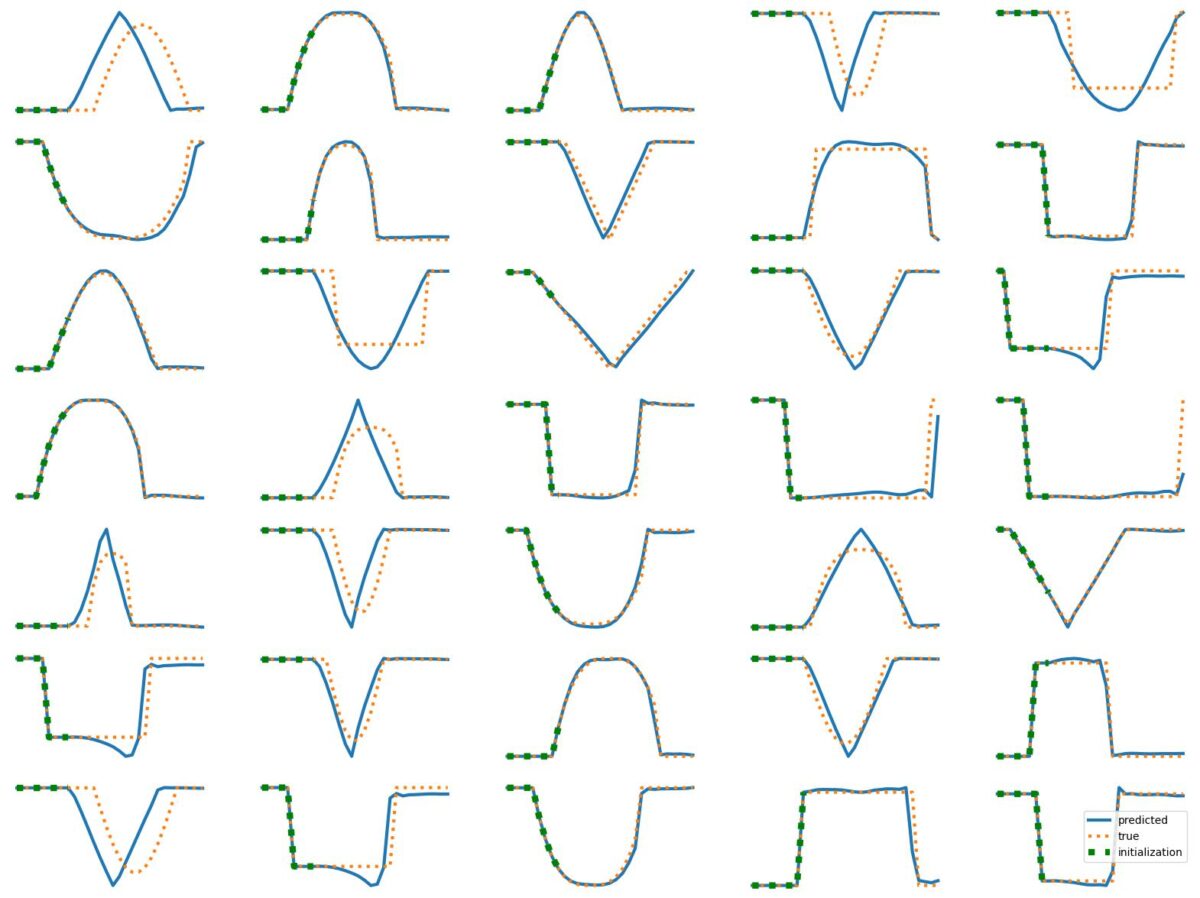
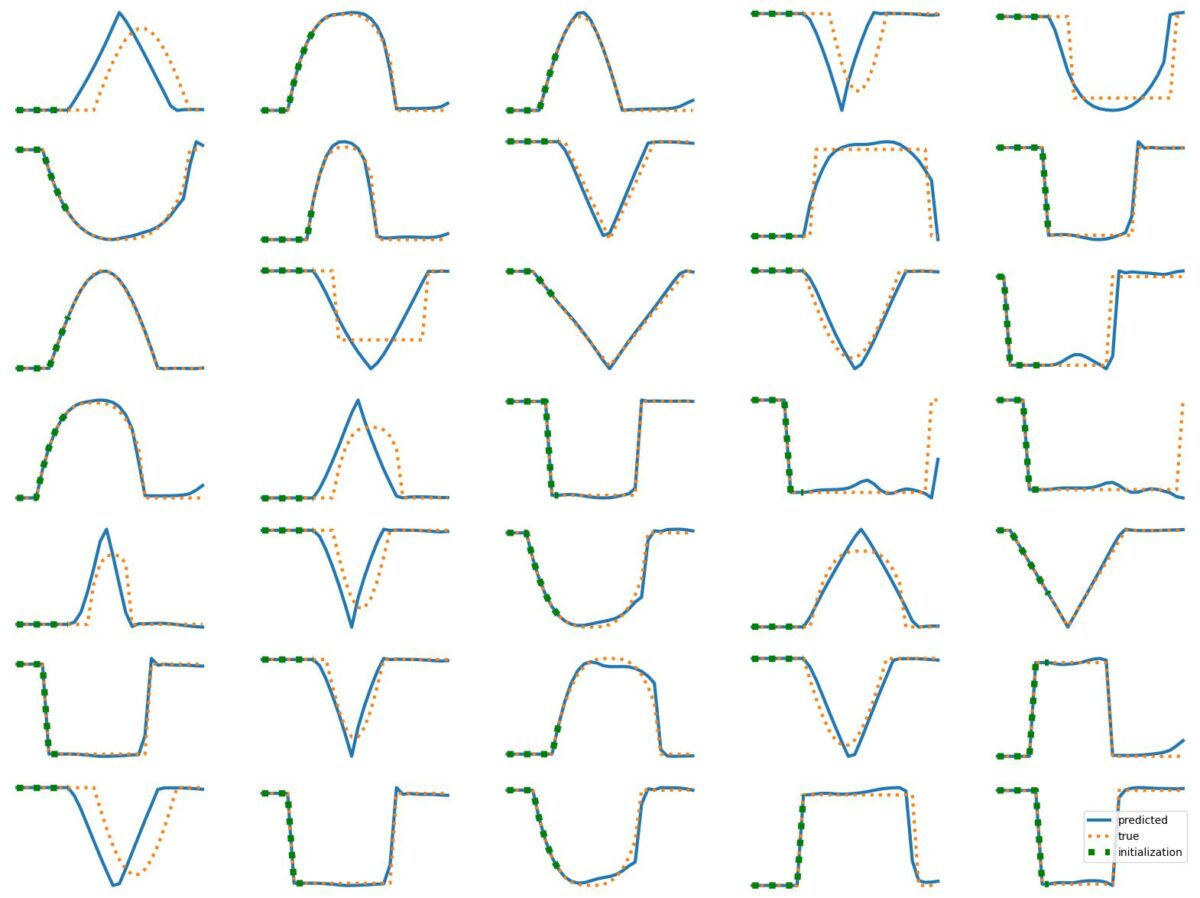
LSTM Examples #1: Basic Time Series Prediction
The results above illustrate how LSTM works with continuous input/output on simple 1D time series prediction tasks. We aim to estimate simple shapes by using past data. We follow an increasingly more complex scenarios. Results show the limitations of continuous time-series prediction via LSTM; the last slides show that even simple shapes cannot be accurately predicted. But the code is helpful to illustrate the basics of time-series prediction.
Below we provide all the commands and the python script needed to generate the slides above.
The commands that generated the slides
# Simplest case
python LSTMtutorial1D.py --data_types=triangle --num_timesteps=1 --use_conv=False
# Simplest case but doesn't work so well if we don't observe sufficient data in given series
python LSTMtutorial1D.py --data_types=triangle --num_timesteps=1 --use_conv=False init_percentage=0.07
# Scale+sign is also no problem
python LSTMtutorial1D.py --data_types=triangle --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True
# we add different types of waves too
python LSTMtutorial1D.py --data_types=triangle,sine --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True
# We even add cube
python LSTMtutorial1D.py --data_types=triangle,sine,cube --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True
# We even add cube, a non-pre-defined shape is predicted particularly when we predict before observing sufficient data
python LSTMtutorial1D.py --data_types=triangle,sine,cube --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True --init_percentage=0.07
# Closely inspect what happens above: What happens when we have only one non-zero entry (i.e., the first value of the wave?) The algorithm doesn't know what to do and completes in a "middle-of-the-way" manner
python LSTMtutorial1D.py --data_types=triangle,sine,cube --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True --num_epochs=50
# Performance somewhat improves when we have use more model parameters
python LSTMtutorial1D.py --data_types=triangle,sine,cube --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True --num_epochs=50 --rnn_hidden_size=100
# We kind of break down when we add the box prediction
python LSTMtutorial1D.py --data_types=triangle,sine,cube,box --num_timesteps=1 --use_conv=False --scale_variation=True --sign_variation=True --num_epochs=100 --rnn_hidden_size=100
# Performance visibly improves when we use two consecutive time frames (it allows the model to capture time derivative, which can uniquely determine the type of shape)
python LSTMtutorial1D.py --data_types=triangle,sine,cube,box --num_timesteps=2 --use_conv=True --scale_variation=True --sign_variation=True --num_epochs=100 --rnn_hidden_size=100
# Performance imrpoves a little more when we use a more complicated model, but still limited
python LSTMtutorial1D.py --data_types=triangle,sine,cube,box --num_timesteps=2 --use_conv=True --scale_variation=True --sign_variation=True --num_epochs=100 --rnn_hidden_size=150
The python script (LSTMtutorial1d) needed for the commands above
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Thu Jul 27 18:34:08 2023
@author: v
"""
import os
import matplotlib.pyplot as plt
import numpy as np
import torch.nn as nn
import torch
import torch.nn.functional as F
import random
import argparse
def str2bool(v):
if isinstance(v, bool):
return v
if v.lower() in ('yes', 'true', 't', 'y', '1'):
return True
elif v.lower() in ('no', 'false', 'f', 'n', '0'):
return False
else:
raise argparse.ArgumentTypeError('Boolean value expected.')
parser = argparse.ArgumentParser(
prog='LSTM-sequence_prediction',
description='Sequence prediction examples via LSTM')
parser.add_argument('--data_types', type=str, default='triangle', required=False,
help="""Which kind of sequences will be used during training & testing?
We have three types: triangle, sine and box. You can use any subset
by separating with commas (e.g., "sine,box" or "triangle,sine,box"")""")
parser.add_argument('--num_timesteps', type=int, default=1, required=False,
help="""How many time steps will be used during preduction?""")
parser.add_argument('--use_conv', default=False, type=str2bool, required=False,
help="""Will convolution layer be added before LSTM? """)
parser.add_argument('--batch_size', default=50, type=int, required=False,
help="""Size of batch (determines num of training samples)""")
parser.add_argument('--num_tra_batches', default=100, type=int, required=False,
help="""Number of batches to use during training (determines """)
parser.add_argument('--num_epochs', default=50, type=int, required=False,
help="""Number of training iterations (i.e., epochs) """)
parser.add_argument('--seq_length', default=30, type=int, required=False,
help="""Length of sequences """)
parser.add_argument('--rnn_hidden_size', default=30, type=int, required=False,
help="Number of nodes in LSTM layer")
parser.add_argument('--scale_variation', default=False, type=str2bool, required=False,
help="Add scale variation to waves")
parser.add_argument('--sign_variation', default=False, type=str2bool, required=False,
help="Add sign variation to waves")
parser.add_argument('--learning_rate', default=0.001, type=float, required=False,
help="Learning rate for optimizer")
parser.add_argument('--init_percentage', default=0.3 , type=float, required=False,
help="Learning rate for optimizer")
args = parser.parse_args()
models_dir = 'models'
if not os.path.exists(models_dir):
os.mkdir(models_dir)
figs_dir = 'figures'
if not os.path.exists(figs_dir):
os.mkdir(figs_dir)
model_path = '%s/Ntra%d-Nep%d-Sc%d-sgn%d-conv%d-Nt%d-T%d-Q%d-%s' % (models_dir, args.num_tra_batches, args.num_epochs, args.sign_variation,
args.use_conv, args.num_timesteps, args.scale_variation, args.seq_length,
args.rnn_hidden_size, args.data_types)
wave_types = args.data_types.split(',')
T = args.seq_length
num_epochs = args.num_epochs
batch_size = args.batch_size
num_tra_batches = args.num_tra_batches
num_tes_batches = 20#args.num_tes_batches
device = 'cuda'
torch.cuda.set_per_process_memory_fraction(0.5, 0)
torch.manual_seed(1907)
random.seed(1907)
def create_single_wave(T, btype = 'sin'):
Ta = round(T/2)
if T % 2 == 0:
Tb = round(T/2)-1
else:
Tb = round(T/2)
if btype == 'triangle':
a = torch.cat((torch.ones(Ta), -torch.ones(Tb)))
x = torch.cumsum(a, 0)
elif btype == 'sine':
x = torch.sin(torch.arange(0,T)*(np.pi/(T-1)))
elif btype == 'box':
x = torch.ones(T)
elif btype == 'cube':
x = (-torch.abs(torch.arange(-Ta, Tb))**3).float()
x = (x-x.min())/(x.max()-x.min())
x = x/torch.norm(x, torch.inf)
return x
def create_naturalistic_signal(T):
Ta = T/3
# acceleration 1, acceleration 2 and acceleration 3
acc1 = create_single_wave(Ta, 'sine')
acc2 = -2*create_single_wave(Ta, 'sine')
acc3 = create_single_wave(Ta, 'sine')
a = np.concatenate((acc1, acc2, acc3), axis=0)
x = np.cumsum(a)
# x = np.cumsum(ca)
return x/np.linalg.norm(x, np.inf)
def generate_shape_sequences(batch_size, wave_types=['triangle'], T = 100, use_diff=False):
Nchannels = 1
if use_diff:
Nchannels += 1
data = torch.zeros(batch_size, T, Nchannels)
for b in range(batch_size):
wave_type = wave_types[random.randint(0, len(wave_types)-1)]
t0 = torch.randint(0, int(T*.40), (1,))+1
tf_0 = T-torch.randint(0, int(T*.40), (1,))
if (tf_0-t0) % 2 == 1:
tf_0 -= 1
tf = tf_0
Tq = tf-t0
Tq = Tq[0].item()
sign = 1
if args.sign_variation:
sign = 1 if torch.rand(1)[0].item() > 0.5 else -1
scale = 1
if args.scale_variation:
scale = torch.rand(1)[0].item()+0.5 # > 0.5 else -1
wave = create_single_wave(Tq, wave_type)*scale*sign
data[b,t0:t0+len(wave), 0] = wave
if use_diff:
data[b,:T-1,1] = data[b,:,0].diff()
data = data.to(device)
return data
class LSTM_seq_prediction(nn.Module):
def __init__(self, input_dim=1, rnn_hidden_size=50, num_layers=1, use_conv=False):
super().__init__()
self.num_layers = num_layers
self.use_conv = use_conv
LSTM_in_channels = input_dim
# print(LSTM_in_channels)
if self.use_conv:
LSTM_in_channels = 4
self.conv = nn.Conv1d(in_channels=1, out_channels=LSTM_in_channels,
kernel_size=2, bias=False)
self.rnn_hidden_size = rnn_hidden_size
self.rnn = nn.LSTM(LSTM_in_channels, rnn_hidden_size,
num_layers=self.num_layers,
batch_first=True)
self.fc = nn.Linear(rnn_hidden_size, 1)
def forward(self, x, hidden, cell):
if self.use_conv:
# Time is typically the second index (after batch_id),
# and we need to move it to last index so that convolution
# is applied over time
x = x.permute(0,2,1)
x = self.conv(x)
x = x.permute(0,2,1)
else:
x = x.permute(0,2,1)
out, (hidden, cell) = self.rnn(x, (hidden, cell))
out = self.fc(out)
return out, hidden, cell
def init_hidden(self, batch_size):
hidden = torch.zeros(self.num_layers, batch_size, self.rnn_hidden_size, device=device)
cell = torch.zeros(self.num_layers, batch_size, self.rnn_hidden_size, device=device)
return hidden, cell
#%%
#
# GENERATE DATA HERE
#
# use_conv = True
# use_diff = False
use_diff = False
# wave_types = ['box', 'triangle', 'sine']
tra_batch_sets = []
tra_batch_label_sets = []
for k in range(num_tra_batches):
batch_data = generate_shape_sequences(batch_size, wave_types=wave_types, T=T, use_diff=use_diff)
tra_batch_sets.append(batch_data.to(device=device))
tes_batch_sets = []
tes_batch_label_sets = []
for k in range(num_tes_batches):
batch_data = generate_shape_sequences(batch_size, wave_types=wave_types, T=T, use_diff=use_diff)
tes_batch_sets.append(batch_data.to(device=device))
#%%
lr = 0.001
model = LSTM_seq_prediction(input_dim=args.num_timesteps,
rnn_hidden_size=args.rnn_hidden_size, num_layers=2,
use_conv=args.use_conv)
model = model.to(device)
loss_fn = nn.MSELoss().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
tra_losses = []
tes_losses = []
t0 = args.num_timesteps
if not os.path.exists(model_path):
for e in range(num_epochs):
closses = []
print(e)
for k in range(num_tra_batches):
hidden, cell = model.init_hidden(batch_size)
data = tra_batch_sets[k]
data = (data-data.mean(axis=1).unsqueeze(1))/data.std(axis=1).unsqueeze(1)
optimizer.zero_grad()
loss = 0
for t in range(t0,data.shape[1]-1):
output, hidden, cell = model(data[:,t-t0+1:t+1,:], hidden, cell)
loss += loss_fn(output[:,:,0], data[:,t+1,0:1])
loss.backward()
optimizer.step()
closses.append(loss.item())
tra_losses.append(np.mean(closses))
with torch.no_grad():
closses = []
for k in range(num_tes_batches):
loss = 0
hidden, cell = model.init_hidden(batch_size)
data = tes_batch_sets[k]
data = (data-data.mean(axis=1).unsqueeze(1))/data.std(axis=1).unsqueeze(1)
for t in range(t0,data.shape[1]-1):
output, hidden, cell = model(data[:,t-t0+1:t+1,:], hidden, cell)
loss += loss_fn(output[:,:,0], data[:,t+1,0:1])
closses.append(loss.item())
tes_losses.append(np.mean(closses))
if (e+1) % int(round(args.num_epochs/3)) == 0 or e == args.num_epochs-1:
plt.subplot(121)
plt.plot(tra_losses)
plt.plot(tes_losses)
plt.subplot(122)
plt.semilogy(tra_losses)
plt.semilogy(tes_losses)
#plt.show()
torch.save(model.state_dict(), model_path)
else:
model.load_state_dict(torch.load(model_path))
#%%
data = tes_batch_sets[4].clone()#.unsqueeze(2)# (batch_size, T, 1)
data = (data-data.mean(axis=1).unsqueeze(1))/data.std(axis=1).unsqueeze(1)
data_orig = data.clone()
M = int(args.init_percentage*T)
# for b in range(batch_size):
# data[b,0:M,0] = torch.arange(M)/(M*10)
for b in range(batch_size):
data[b,M:,:] = 0 # wave[0:M]#/(M*10)
# for b in range(batch_size):
# data[b,M-1:M,0] = 1 # torch.arange(M)/(M*10)
# data = (data-mu)/std
# data_orig = (data_orig-mu)/std
with torch.no_grad():
closses = []
hidden, cell = model.init_hidden(batch_size)
for t in range(t0,M):
output, hidden, cell = model(data[:,t-t0:t,:], hidden, cell)
for t in range(M,T):
output, hidden, cell = model(data[:,t-t0:t,:], hidden, cell)
data[:,t:t+1,0:1] = output[:,:,0:1]
data[:,t:t+1,1:2] = 1*(data[:,t:t+1,0:1]-data[:,t-1:t,0:1])
lw = 3
plt.figure(figsize=(20, 15))
for b in range(35):
plt.subplot(7, 5, b+1)
plt.plot(data[b,:,0].to('cpu').squeeze(), linewidth=lw)
plt.plot(data_orig[b,:,0].to('cpu').squeeze(), ':', linewidth=lw)
plt.plot(data_orig[b,:M,0].to('cpu').squeeze(), 'g:', linewidth=lw*2)
plt.axis('off')
plt.legend(['predicted', 'true', 'initialization'])
fig_path = '%s/Ntra%d-Nep%d-Sc%d-sgn%d-conv%d-Nt%d-T%d-Q%d-%s-%.3f.jpg' % (figs_dir, args.num_tra_batches, args.num_epochs, args.sign_variation,
args.use_conv, args.num_timesteps, args.scale_variation, args.seq_length,
args.rnn_hidden_size, args.data_types, args.init_percentage)
plt.savefig(fig_path,bbox_inches='tight')
# plt.show()
# plt.show()
if args.use_conv and args.num_timesteps == 2:
print('Convolution parameters are')
print('==========================')
print(next(iter(model.conv.parameters())))